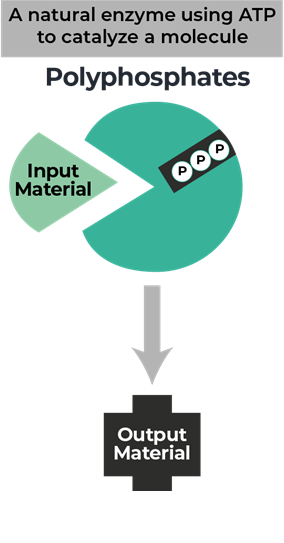
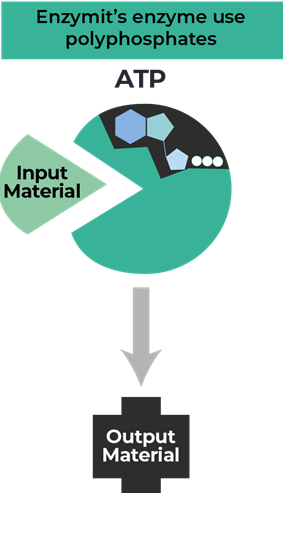
Resource Center
Case Studies
Novel Serum Proteins to Catalyze Cultivated Meat Production
One of the major challenges of the cultivated meat industry is the cost of growth factors, unique protein molecules that stimulate the growth and maintain the health of cell tissue. Growth factors play an essential role in promoting cellular differentiation and cell division. However, deriving from mammalian cells, they generally cannot be produced through microbial fermentation, resulting in unfeasible production expenses that hinder cost reduction and customer adoption.
To address the financial and developmental challenges of cost-effective insulin manufacturing, Enzymit has designed multiple novel insulin-like proteins, a crucial growth factor in cell growth, but also an important therapeutic. These new molecules, some with sequence identity completely different from native insulin, can be expressed in bacterial cells, making large-scale production significantly more cost-effective.
Biosensors utilize proteins that can sense and detect the presence of particular substances, or measure specific properties of a sample, for a range of applications. These may include environmental monitoring, safety monitoring, and industrial process control.
Novel biosensors for ground contaminants may be driven by a need to monitor and control the presence of harmful substances to protect public health and safety, or to ensure compliance with regulatory standards. They offer several advantages over traditional sensors, such as higher sensitivity, greater specificity, and more rapid response times, providing an ideal solution for detecting and measuring contaminants in the ground.
Enzymit is designing multiple novel biosensors that can identify trace amounts of harmful substances with far higher specificity and minimal signal-to-noise ratio compared to traditional detection methods.
There i currently no commercially available enzyme for polyolefin degradation, and traditional chemical degradation methods are expensive and can only be used for a limited time.
Enzymit is developing novel enzymatic solutions that address the specific challenges of plastic degradation to help realize a more sustainable and environmentally responsible future.

There i currently no commercially available enzyme for polyolefin degradation, and traditional chemical degradation methods are expensive and can only be used for a limited time.
Enzymit is developing novel enzymatic solutions that address the specific challenges of plastic degradation to help realize a more sustainable and environmentally responsible future.

The foundation of modern society is hydrocarbons, mostly obtained from fossil fuels. However, as these resources are becoming scarcer and their excessive Use negatively impacts the environment. Drop-in biofuels, which provide functionality to traditional fuels without the environmental drawbacks of earlier-generation biofuels (such as ethanol and biodiesel) are proving to be a more viable solution.
The current technology for drop-in biofuel production relies on chemical hydrogenation, which requires vast investments in infrastructure and limits geographical distribution. Enzymit’s technology allows us to build smaller factories that can be erected faster and more cost-effectively where they are needed, dramatically increasing the market potential.
Zimmerman L, et al. Context-Dependent Design of Induced-Fit Enzymes Using Deep Learning Generates Well Expressed, Thermally Stable and Active Enzymes.” BioRxiv, Aug. 2023
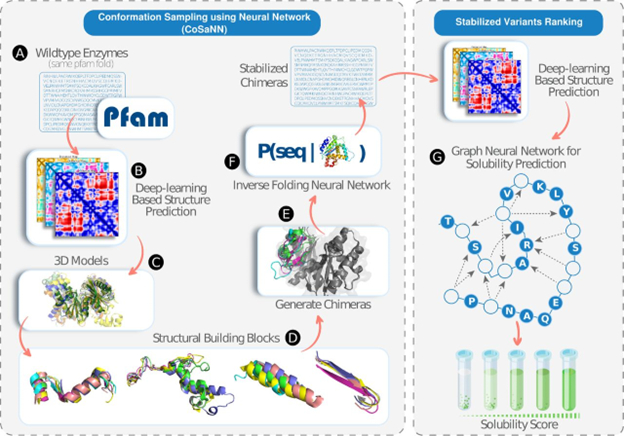
Abstract
The potential of engineered enzymes in practical applications is often constrained by limitations in their expression levels, thermal stability, and the diversity and magnitude of catalytic activities. De-novo enzyme design, though exciting, is challenged by the complex nature of enzymatic catalysis. An alternative promising approach involves expanding the capabilities of existing natural enzymes to enable functionality across new substrates and operational parameters. To this end we introduce CoSaNN (Conformation Sampling using Neural Network), a novel strategy for enzyme design that utilizes advances in deep learning for structure prediction and sequence optimization. By controlling enzyme conformations, we can expand the chemical space beyond the reach of simple mutagenesis. CoSaNN uses a context-dependent approach that accurately generates novel enzyme designs by considering non-linear relationships in both sequence and structure space. Additionally, we have further developed SolvIT, a graph neural network trained to predict protein solubility in E.Coli, as an additional optimization layer for producing highly expressed enzymes. Through this approach, we have engineered novel enzymes exhibiting superior expression levels, with 54% of our designs expressed in E.Coli, and increased thermal stability with more than 30% of our designs having a higher Tm than the template enzyme. Furthermore, our research underscores the transformative potential of AI in protein design, adeptly capturing high order interactions and preserving allosteric mechanisms in extensively modified enzymes. These advancements pave the way for the creation of diverse, functional, and robust enzymes, thereby opening new avenues for targeted biotechnological applications.
David L, et-al. Performance Upgrade of a Microbial Explosives’ Sensor Strain by Screening a High Throughput Saturation Library of a Transcriptional Regulator.” Computational and Structural Biotechnology Journal, vol. 21, 2023.
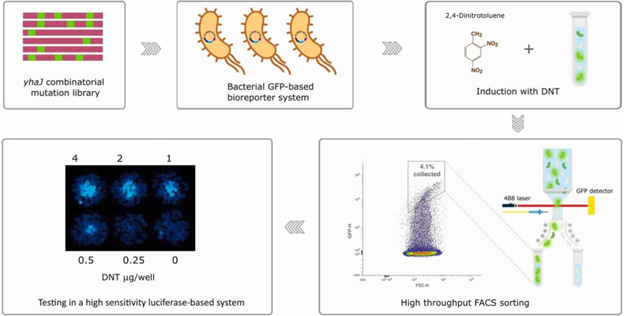
Abstract
We present a methodology for a high-throughput screening (HTS) of transcription factor libraries, based on bacterial cells and GFP fluorescence. The method is demonstrated on the Escherichia coli LysR-type transcriptional regulator YhaJ, a key element in 2,4-dinitrotuluene (DNT) detection by bacterial explosives’ sensor strains. Enhancing the performance characteristics of the YhaJ transcription factor is essential for future standoff detection of buried landmines. However, conventional directed evolution methods for modifying YhaJ are limited in scope, due to the vast sequence space and the absence of efficient screening methods to select optimal transcription factor mutants. To overcome this limitation, we have constructed a focused saturation library of ca. 6.4 × 107 yhaJ variants, and have screened over 70 % of its sequence space using fluorescence-activated cell sorting (FACS). Through this screening process, we have identified YhaJ mutants exhibiting superior fluorescence responses to DNT, which were then effectively transformed into a bioluminescence-based DNT detection system. The best modified DNT reporter strain demonstrated a 7-fold lower DNT detection threshold, a 45-fold increased signal intensity, and a 40 % shorter response time compared to the parental bioreporter. The FACS-based HTS approach presented here may hold a potential for future molecular enhancement of other sensing and catalytic bioreactions.